Proximity labeling works by fusing the decoy protein to be studied with an enzyme that nonspecifically labels the surrounding protein. The tagged proteins are called "prey" proteins.
There is a rapid development of protein labeling techniques based on proximity-tagging enzymes. Biotin-dependent biotin identification (BioID) of biotin ligase mutants relying on biotin ligase (BirA), which can label lysine residues of neighboring proteins with biotin tags in the presence of biotin. Biotin labeling relies on engineered ascorbate peroxidase (APEX), which enables rapid biotinylation in the presence of hydrogen peroxide in a short period of time, facilitating the study of protein interaction networks in space and time. PUP-IT, a proximity labeling technology relying on the PUP ligase PafA. AirID, a new proximity marker enzyme made by targeted mutation of BirA from metagenomic data. RaPID for studying RNA-protein interactions, and CRUIS protein proximity tagging technology that combines CRISPR and PUP-IT, etc.
These methods require less stringent pull-down conditions than those required by traditional methods. They are also not required to remain bound when analyzing protein complexes and can tolerate more intense lysis conditions. It allows labeling of proteins in cells under normal physiological conditions without mismatching interacting proteins in the lysis and affinity purification steps, reducing false positive results. It has been successfully applied in mammalian cells, plants, parasites, mucor, mice, yeast, zebrafish, Drosophila and worms.
BioID
BioID can label proteins that are physically spatially proximate to decoy proteins with biotin as evidence of protein interactions.
A recombinant expression vector of the target protein and BirA is constructed and transfected into cells for amplification and expression. Afterwards, biotin is added to the medium of the transfected cells. Since BirA can activate biotin, all relevant proteins within 10 nm diameter around the target protein can be labeled by biotin. The biotinylated proteins were enriched from the samples by streptavidin-coated magnetic beads and then analyzed by mass spectrometry identification.

BioID workflow for the identification of direct and proximal protein-protein interactions (Cheerathodi et al., 2020)
Learn more: dynamic light scattering protein
Split-BioID
Split-BioID is a protein fragment complementation analysis method based on the BirA* (mutation at position 118 of BirA) enzyme that allows spatial and temporal analysis of protein complexes.
Split-BioID works by splitting BirA* into two fragments that are attached to two interacting proteins. Upon fusion of the two interacting proteins together, BirA* can be reassembled to restore activity.
When applying this method, a suitable splitting site can be selected experimentally. A common method fuses the split BirA* to the binding site of the FKBP12-rapamycin complex (FRB) and the FK506 binding protein (FKBP), respectively. Upon addition of rapamycin, FRB can interact with FKBP to rejoin the two fragments of BirA*, thus allowing the recovery of its biotinylation activity to be verified.

Design of split-BioID (Schopp et al., 2017)
BioID can label in vivo proteins of interest in the 10 nm range with biotin, regardless of the interaction. In contrast, Split-BioID is activated in the natural cellular environment only when two proteins interact with each other. The ability to unbiasedly detect complexes formed by a pair of interacting proteins in a single and simple analysis makes this method unique among PPI analytical methods.
Contact-ID
Contact-ID is also a type of split-BioID. BirA* was divided into two fragments, N-G78 (B1) and G79-C (B2). These two fragments are fused to the SEC61B nitrogen terminus (B1-SEC61B) located in the ER membrane (ERM) and the TOM20 carbon terminus (TOM20-B2) located in the outer membrane of the mitochondrial membrane (OMM), respectively. The biotinylation activity is dependent on the formation of mitochondria-associated membranes (MAMs). Biotinylation capacity is restored only when they physically interact and reorganize at the membrane contact site.

Schematic view of the restoration of biotinylating activity of split-BioIDs using the FRB–FKBP system in the presence of rapamycin (Kwak et al., 2020)
When labeling the proteome of two cell membrane contact sites in situ, the following two conditions must be met:
(1) The biotinylation activity of the Split-BioID system is restored only after the proximity of the two fragments and can be verified by proximity to a controlled biological system, such as rapamycin-induced protein-protein interactions;
(2) The intrinsic affinity between the two split fragments should be negligible to avoid artificial MAM formation and subsequent false-positive identification.
The method specifically biotinylates proteins localized to the MAM and identifies the biotinylated sites by mass spectrometry, thus unambiguously revealing the MAM proteome in living cells.
BioID2
BioID2 (231 AA), a biotin ligase R4OG mutant derived from A. aeolicus, naturally lacks the DNA-binding structural domain of biotin ligase and is approximately one-third smaller than BioID. The reduced volume enhances the targeting and localization of the protein fused to it.

The dimensions of E. coli (left; PDB ID 1BIA) and A. aeolicus (right; PDB ID 2EAY) biotin ligases based on prior structural analyses (Kim et al., 2016)
BioID2 requires lower biotin concentration and shorter time to induce biotinylation compared to BioID. This property facilitates its use in systems where biotin replenishment is difficult. The optimal reaction temperature of BioID2 is about 50 °C (the optimal temperature of BioID is about 37 °C), which is suitable for labeling of thermophilic bacteria.
When the target protein is significantly larger than the labeling radius of BioID2 or when the fusion protein is a larger protein complex, its labeling range can be extended by adding flexible linker chains.
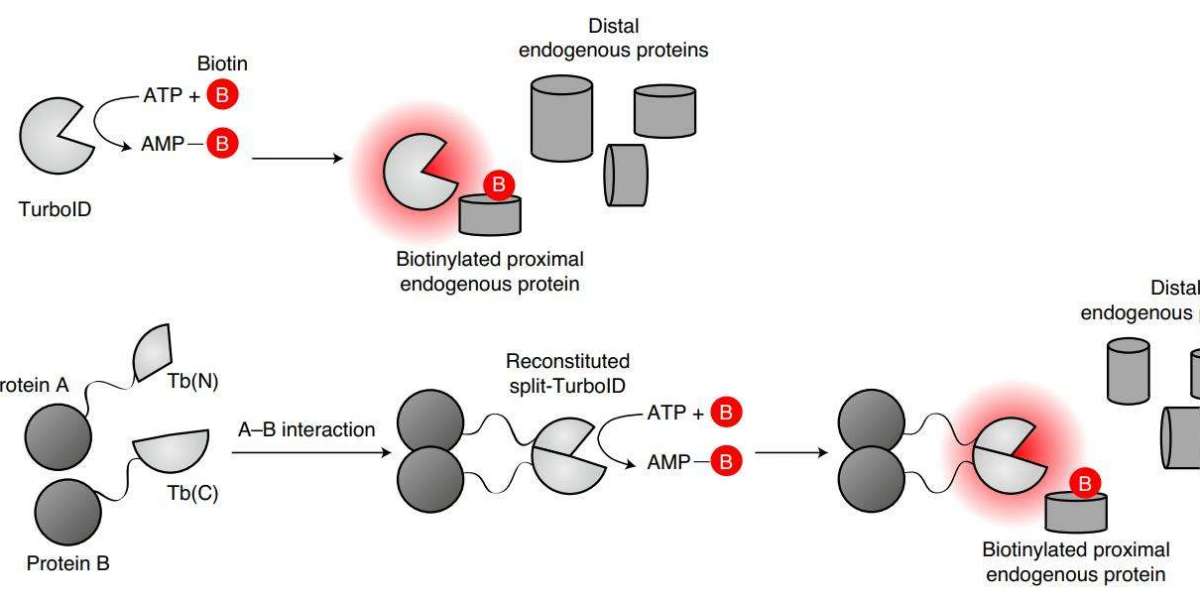
TurboID & miniTurbo
TurboID and miniTurbo were generated by R118S mutation of BirA by PCR with high error rate and screened by expressing it on the yeast surface. Their catalytic efficiency is higher than that of BirA. They can act as neighboring markers within 10 min of adding biotin.
TurboID is also 35 kD in size and has 15 mutations compared to wild-type BirA. miniTurbo is 28 kD in size, lacks the N-terminal structure, and has 13 mutations compared to wild-type BirA. Although the biotinylation activity of miniTurbo is lower than that of TurboID, its less labeling and clean background before the addition of exogenous biotin make miniTurbo a better choice in experiments that require precise control of labeling time.
Split-TurboID is similar to split-BioID. TurboID is divided into two inactive fragments with high recombination capacity and ligated to FRB-FKBP. It is then fused to proteins expressed in different subcellular compartments separately to establish biotin- and rapamycin-dependent expression systems. The recombination of the two components of TurboID is activated when protein-protein interactions or membrane-membrane contacts occur, achieving proximity labeling.

Proximity-dependent biotinylation catalyzed by TurboID and split-TurboID (Cho et al., 2020)
May et al. established cell lines stably expressing BioID and TurboID fusion proteins and compared the two by immunoblotting, immunofluorescence and biotin affinity-based purification proteomics. TurboID was found to show signs of protein instability. Sustained biotinylation was also possible in the absence of exogenous biotin, and the actual labeling radius was increased. However, the biotinylation of TurboID in the endoplasmic reticulum lumen was more potent than that of BioID.
AirID
AirID was first reported in 2020 and was obtained by Kido et al. from their own library (a library with 30% homology to E. coli BirA built by Blastp web server and custom Python script). The proximal biotinylation activity of BirA* is significantly lower than that of TurboID and AirID. AirID requires a lower amount of biotin for labeling than TurbolD (TurboID requires 50 umol/L biotin, while AirID can achieve proximal labeling in a 5 umolL biotin or even biotin-free system). Biotinylation of AirID occurs on adjacent lysine residues and has no specific sequence preference, and the high accuracy of biotinylation can also be used to detect the inhibition of PPI by drugs.
References
- Cheerathodi, M. R., & Meckes, D. G. (2020). BioID combined with mass spectrometry to study herpesvirus protein–protein interaction networks. In Herpes Simplex Virus (pp. 327-341). Humana, New York, NY.
- Schopp, I. M., Amaya Ramirez, et al. (2017). Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nature communications, 8(1), 1-14.
- Kwak, C., Shin, S., et al. (2020). Contact-ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial-associated membrane formation. Proceedings of the National Academy of Sciences, 117(22), 12109-12120.
- Kim, D. I., Jensen, S. C., et a. (2016). An improved smaller biotin ligase for BioID proximity labeling. Molecular biology of the cell, 27(8), 1188-1196.
- Cho, K. F., Branon, T. C., et a. (2020). Proximity labeling in mammalian cells with TurboID and split-TurboID. Nature Protocols, 15(12), 3971-3999.